Analyte Identification
The accurate mass measurement of an analyte is used to calculate possible molecular formulae that are closest to the observed monoisotopic mass. Frequently several candidate formulae are possible within the margin of error (typically ± 5 ppm for data from LC or GC TOF instruments) that preclude a definitive assignment. The isotopic pattern of the analyte's spectrum (spacing and relative intensities) also provides valuable information regarding it's elemental composition. The presence of heteroatoms with distinct isotopic abundancies, e.g. Cl, Br, Fe etc., can often be deduced in organic compounds. Furthermore, the 13C isotopic peak abundance also gives a good estimate of the # of carbons present.
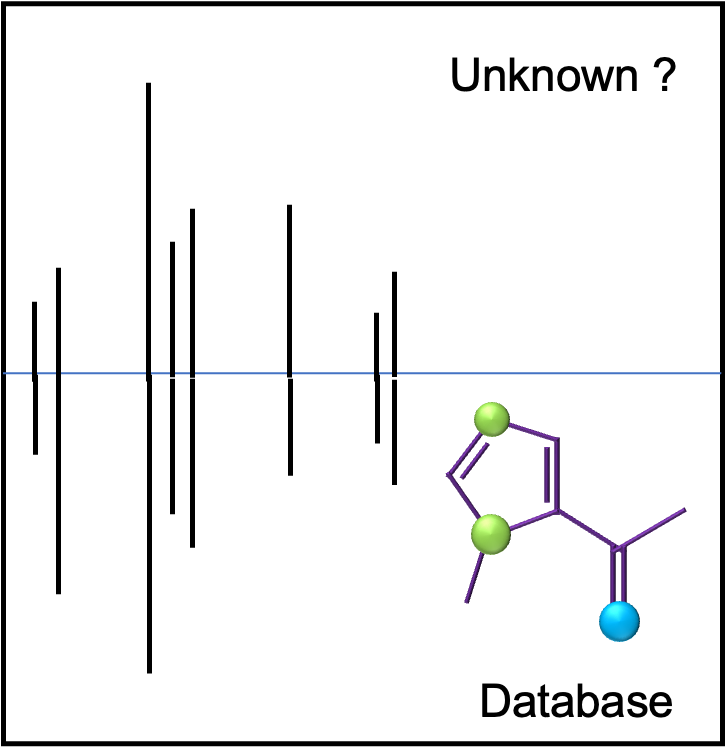
Additional information from a fragmentation spectrum of the analyte is needed to increase the confidence in an identification. Hard ionization techniques (EI and CI) used in GC-MS techniques give reproducible fragmentation spectra suitable for database searching and matching. High-quality compound spectral libraries, such as the NIST GC EI spectral library, contain hundreds of thousands of entries (NIST 2020: 350,000 EI Spectra; 447,000 retention index (RI) values). Library search results are scored based on overlap in # of peaks present and their relative peak intensities. If a compound is represented in the library, a good spectral match should be found.
Fragmentation spectra from LC-MS2 data are also informative but are more variable. Fragmentation patterns from soft ionization techniques (e.g. electrospray ionization) can be instrument specific and/or dependent on specific ionization parameters and LC conditions. Experimentally derived LC-MS2 libraries (e.g. METLIN) are also not as complete. Computational approaches to augment libraries by generation of theoretical fragmentation spectra are improving but have limits. Consequently, identification of an analyte may not be positively confirmed without additional experimental analyses (NMR, co-elution studies etc.).
Analyte identifications can be challenging and time consuming, especially for complex mixtures such as biological matrices (tissue extracts, environmental samples) that can contain thousands of analyte features many of which may co-elute or are present in highly variable abundance.
Staff are available to assist and guide in data interpretation. The NIST library is integrated into software on all the GC-MS instruments. Additional links to other searchable open source databases are available.